SOLIDA
To facilitate the reproducibility and portability of NGS pipelines, we have develop SOLIDA, a Python command-line tool that can easily organize the deployment, the data management and the execution of a Snakemake-based workflow.
What is SOLIDA?
SOLIDA is a tool that can easily bootstrap any Snakemake workflow organizing its activity in different properly configured projects.
Given that each project can differ for pipeline code, input data, workflow configuration, virtual environment and/or working folder, SOLIDA is able to manage all these tasks in a very simple way.
Why should I need SOLIDA?
| Download | Setup | Deploy |
|---|---|---|
 |
 |
 |
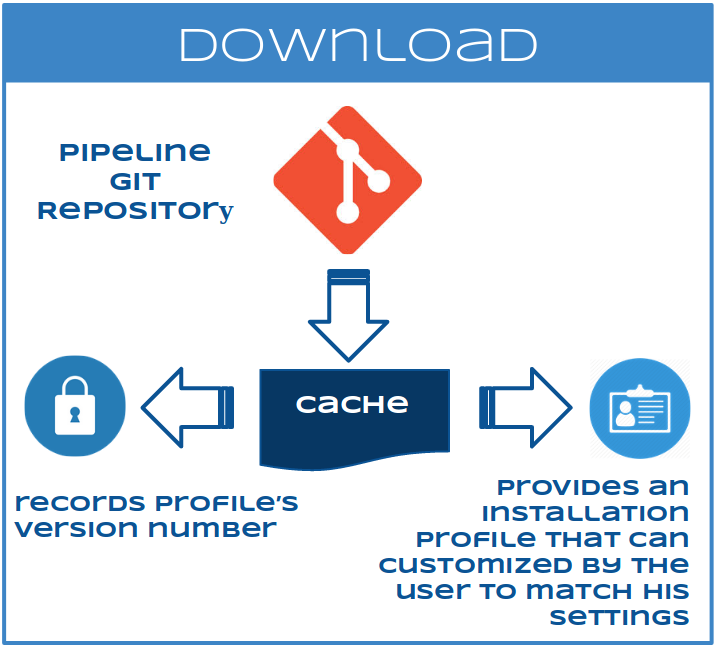
SOLIDA downloads each pipeline (e.g. exome, RNA-Seq, smallRNA-Seq) from a git repository into a cache store, records its version number and provides an installation profile that can be easily customized by the user to match his settings.
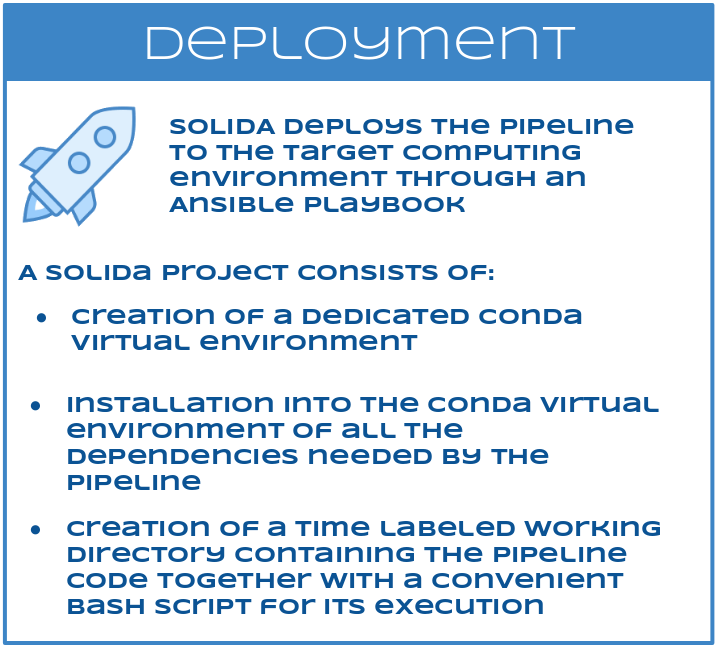
SOLIDA then deploys, for each project, the pipeline to the target computing environment through an Ansible playbook, as follows:
- creation of a dedicated Conda virtual environment
- installation into the Conda virtual environment of all the dependencies needed by the pipeline
- creation of a time labeled working directory containing the pipeline code together with a convenient bash script for its execution
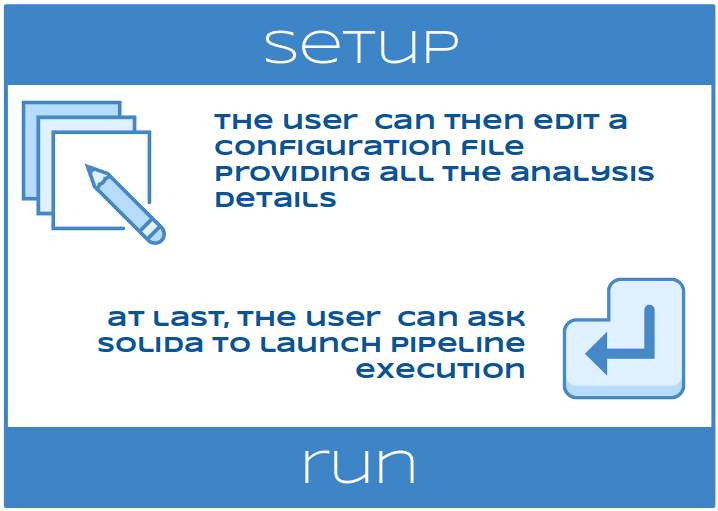
The user can then edit a configuration file providing all the analysis details and ask SOLIDA to launch its execution. For users convenience, a set of different “ready to go” pipelines to analyze DNA-Seq (exome and targeted resequencing), RNA-Seq (reference based and pseudo-alignments) and smallRNA-Seq data have been included in SOLIDA, but others can be added quickly.
Download and Install
You can install the latest stable version of SOLIDA from PyPI:
pip install solida
or you can clone the GitHub repository and install it:
git clone https://github.com/solida-core/solida.git
cd solida
python setup.py install
Requirements
To run Solida, Conda must be present in your computer. To install it, see https://conda.io/miniconda.html
Usage
To check version:
solida -v
To list all the pipelines enabled, digit
solida info
To check if both pipeline and profile are available, digit:
solida setup -l pipeline_label -p profile_label
Before to deploy a pipeline, you have to create a project profile:
solida setup -l pipeline_label -p profile_label --create-profile
Solida will create a yaml file named profile_label.yaml into ~/solida_profiles Edit the profile_label.yaml to match your environment settings.
After that, deploy the pipeline into localhost with:
solida setup -l pipeline_label -p profile_label --deploy
If you want to deploy the pipeline into a remote host, add these arguments:
solida setup -l pipeline_label -p profile_label --deploy --host remote_host
--remote-user username --connection ssh
where: –host is the hostname of the remote host –remote-user is a username available in the remote host –connection is the type of connection to use
Pay attention: remote_user have to be able to do ssh login into remote_host without password (SSH Key-Based Authentication)
Script to execute the workflow

NEW FEATURE !
We recently released a Django-based Graphical Interface for SOLIDA. It comes within a Docker container which can be cloned from solida-core/solida-gui GitHub page.

